Molecular Phylogeny
Molecular phylogeny is a method used to analyze molecular differences in heritable traits within a gene. These traits are compared to other organisms that are thought to be genetically similar on order to determine whether there is an evolutionary relationship between them. Phylogenetic trees can give us a bigger picture of the relationships between species, if they have common ancestors, or if the species or phylogenic groups can be traced back to in the phylogenic tree to a point where we can hypothesize a general time frame when one subset of a species genetically diverged from an original. It will be important to remember that a species includes organisms able to interbreed and produce viable offspring. When imagining the origins of life, we can see how putting together all evolutionary relationships would form a tree, with the most shared common ancestors at the base and trunk, and the most recent species forming the branches. Below is an example of a phylogenetic tree retrieved from Chapter 23 of Launchpad:
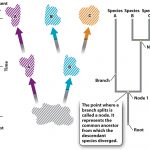
Looking above, one can see that the origin of the tree in question is called the “root” named so because that is the most common ancestor of species A, B, and C. the forks where we can see genetic divergence is called a node. The lines coming off of that node is called a Branch, which can be followed to yet another divergent species. The more branches observed in a phylogenetic diagram, the more speciation that occurred.
References
Biology: How life works (2nd ed.). (2017). In B. A. Berry Andrew, Biology: How life works (2nd ed.) (p. CH 23). McGraw Hill, NY: W. H. Freeman. Retrieved from http://www.macmillanhighered.com/launchpad/morris2e/4909413#/ebook/item/MODULE_bsi__F4951CED__2971__4486__BC4F__E109B2EE87D4/bsi__2ED633B9__F1E8__4C6C__82AA__0633C0C9F474?mode=Preview&toc=syllabusfilter&readOnly=False&renderIn=fne